Luminous shrimp allows for faster detection of infectious diseases
Professor Maarten Merkx on a more widely applicable PCR alternative
Read more- Research
- 14/02/2024
Luminous shrimp allows for faster detection of infectious diseases
The PCR test was the gold standard during the Covid-19 pandemic. The waves of Covid may have subsided now, but should another pandemic break out, the research group of TU/e professor Maarten Merkx is working hard on a faster and cheaper alternative. The key ingredient in this method: a luminous shrimp.
Many of us still shudder at the memory of having a cotton swab stuck up our nose. “Would you prefer your left or your right nostril?” was a common question at the numerous Covid testing sites. But what exactly does a PCR test do?
Multiplying genetic material
“PCR (polymerase chain reaction, Ed.) is a method by which many copies can be made from just one DNA molecule,” explains Maarten Merkx, professor of Protein Engineering at the Department of Biomedical Engineering. If you are infected, your nasal fluid on the swab contains genetic material from the virus, in the form of DNA or RNA.
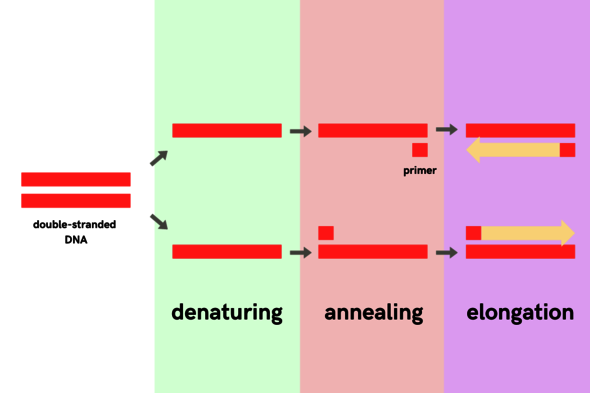
The amount of genetic material on such a swab is often minimal. So minimal, in fact, that no reliable test can be carried out to determine whether you are infected or not. PCR is a method used to multiply a specific portion from small amounts of genetic material (such as on a swab) over and over again. This continues until there is enough present to detect it. This copying process involves three steps that are repeated many times. First, the two DNA strands are “melted” apart at high temperatures, then at lower temperatures, the specific DNA sequence is delineated and copied.
A PCR test for Covid-19 multiplies a specific sequence of the coronavirus RNA, which allows you to determine from the test whether or not it is present and thus, whether or not the person is infected.
The three steps of PCR
Preparation
PCR consists of three steps: denaturation, annealing and elongation. To start the process, a DNA template is required. The genetic material of the coronavirus consists of RNA. This is similar to DNA but consists of a single strand rather than a double strand. In addition, RNA has slightly different building blocks than DNA. By treating it with the “reverse transcriptase” enzyme, the RNA is converted into DNA; and then the PCR can begin.
Step 1: Denaturation
The DNA sample is heated to about 95 degrees Celsius. Because of the high temperatures, the two coiled strands “melt” apart.
Step 2: Annealing
The primers, which function as flags delineating the piece of DNA you are looking for, now have room to bind to the DNA. In the case of a PCR test for Covid, the primers bind to a stretch of genetic material specific to the coronavirus. So if this genetic material is not present (because the person in question does not have Covid-19) then the primers will not be able to bind. In this step, the sample is cooled down to about 60 degrees Celsius because the primers are most effective at that temperature.
Step 3: Elongation
Now, the copying process can begin. The enzyme responsible for this is called DNA polymerase; it reads the area between the two primers and makes a copy of it. If the primers were unable to bind, because there is no coronavirus present, nothing will be copied either.
A full day for your results
“In order to obtain enough DNA to be able to measure this, you have to perform such a reaction 30 to 40 times. One reaction takes a few minutes, which means a complete PCR reaction takes about two hours,” Merkx says.
However, during the Covid-19 pandemic, it generally took a full day, or sometimes even two, before you could expect your results. In addition to the duration of the testing process, Merkx says the required PCR equipment was also a bottleneck. “PCR requires specific equipment (for the temperature control, Ed.), which you can really only find in a medical microbiology laboratory and not at the GP, for example,” he says. So the need for specific equipment, qualified staff, and travel time from the sample collection site to the laboratories also played a role.
And then there was the self-test, of course. Again, a swab up your nose, but this time from the comfort of your own home, at the dining table. “This is used to measure the presence of proteins from the virus instead of DNA or RNA,” explains Merkx. “They are fast, and you can do them at home, but they’re not as sensitive.” So they are useful in order to get a general idea, but due to their low sensitivity, they are no substitute for the PCR test. “The essence of our new method is that it is as fast as the self-test, but as sensitive as a PCR test,” says Merkx.
The essence of our new method is that it is as fast as the self-test, but as sensitive as a PCR test
Your results as you wait
There is a reason why Merkx aims for both speed and sensitivity. Ultimately, he hopes to introduce his method to general practitioners and other health care providers. “The big advantage would be that you get your results immediately during your visit to the GP. This way you instantly know whether or not you need to stay at home, or whether or not you need medication.” To realize this, two alternatives had to be found.
Firstly, an alternative to PCR. A method to multiply genetic material, like PCR, but faster, easier to perform, and requiring less complex equipment.
Secondly, an alternative for fluorescence was needed.“With PCR, you have the copying process, of course, but then also a way to detect the copied DNA. This is often based on fluorescence,” explains Harm van der Veer, PhD candidate in Merkx’s research group. Fluorescence makes the copied target DNA (e.g., from the coronavirus) light up when it is present. This is how the people in the lab know whether or not the person is infected. “This also requires a device that you don’t typically find at the GP’s office,” Van der Veer adds. Therefore, they set out to find a luminous alternative that would also work without elaborate equipment.
The big advantage would be that you get your results immediately during your visit to the GP
RPA instead of PCR
To speed up and simplify the copying process, they opted for RPA (recombinase polymerase amplification), a method that replicates genetic material just like PCR. Unlike PCR, you don’t have to continuously change the temperature for this. Instead of melting, we use an enzyme that basically pushes the two parts of a DNA strand apart at a low temperature,” explains Van der Veer. “The entire process can take place at that single low temperature.” According to Merkx and Van der Veer, not having to change temperatures, as is currently required for PCR, has significant advantages.
Firstly, the equipment is less complex and therefore less expensive. Secondly, the copying step takes only 10-15 minutes because no temperature changes are required after each step, making it possible to immediately copy a newly formed DNA molecule. “The result is a much faster exponential increase (of genetic material, Ed.), Merkx explains. “That’s critical if you need to do a test quickly.”
Bioluminescence instead of fluorescence
“This (RPA, Ed.) is already being used, but when it comes to reading out the results, you’re often bound to fluorescence, just like with PCR,” says Van der Veer. “That’s where our bioluminescence readout method comes into play.”
Bioluminescence is the production of light by organisms. The most famous example is fireflies, but various bacteria and many deep-sea creatures are also capable of generating light themselves. “About 50 percent of deep-sea organisms can produce light,” says Merkx. This actually makes perfect sense, because it is pitch dark deep down in the sea.
The major advantage of bioluminescence over fluorescence is that it is easier to see. If it is bright enough, it is even visible to the naked eye. However, analyzing it more accurately requires a digital camera – like the one on your smartphone.
luciferine + O₂ + luciferase —> oxyluciferine + CO₂ + licht
Luminous shrimp
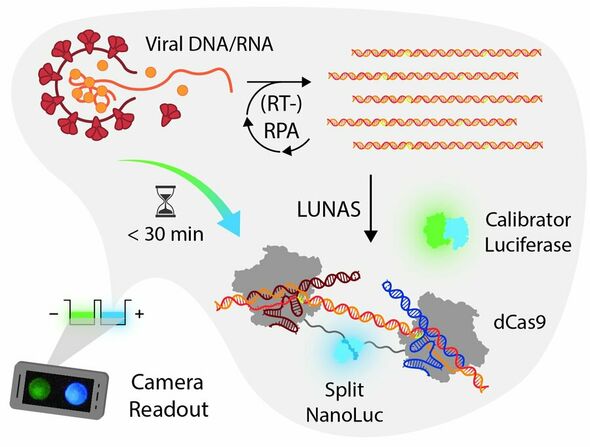
Merkx’s bioluminescence readout method is called LUNAS (LUminescent Nucleic Acid Sensor) and uses the luminous enzyme NanoLuc, derived from a shrimp that lives kilometers deep in the sea (official name: Oplophorus gracilirostris). This is what allows the shrimp to produce light on its own. “They use this (light, Ed.) as a distraction”, Merkx explains. “When they’re in danger, for example, because they’re about to be eaten by another animal.”
But how does the shrimp produce this light? “The shrimp simultaneously ejects both the enzyme, luciferase (NanoLuc, Ed.), and the substrate molecule (otherwise known as luciferin, Ed.). That substrate molecule is oxidized by the enzyme,” says Van der Veer. “During this process, a new product molecule is created, as well as a photon. And that’s what emits the light.”
Every bioluminescent species has a specific kind of luciferase, which determines, among other things, the color of the light produced. In the case of the sea shrimp in question, the light is blue.
In practice
But how does this work in practice? The general practitioner or another healthcare provider takes your sample. Using RPA, the DNA/RNA from your sample is copied until there is enough to be detected.
To ensure that the blue light reaction only occurs when the target DNA/RNA (for example, from the coronavirus) is present, the blue luciferase from the shrimp (NanoLuc) is split into two parts. Each part is linked to a protein that can detect the target DNA/RNA (the CRISPR protein called dCas9).
If the target DNA/RNA is present, then one of the dCas9 proteins (with the first NanoLuc fragment) binds at a specific location, and the other dCas9 protein (with the second NanoLuc fragment) binds right next to it. This brings the two halves of NanoLuc together so that they can become one again, which results in the production of blue light. If the target DNA/RNA is not present, the dCas9 proteins will not be able to bind to anything, leaving the luciferase split and thus unable to produce blue light.
“The test always produces green light,” Van der Veer says. This involves a second luciferase, the calibrator, and serves as a control. “We linked this second luciferase to a green fluorescent protein, resulting in the production of green light,” Merkx explains. “If the target DNA is present, it changes from green to blue light and then it stays blue. In order to determine the result, whether someone is infected or not, we measure the ratio of green and blue light,” Merkx says.
This complete process, from RPA to the color change, takes about 15 minutes and can be carried out with simple equipment. “We’ve proven that you can read it out well using a digital camera, like the one on your smartphone.” So it is ideal for use at the general practitioner’s office.
Does this replace the PCR test?
Yes and no. “Our method is a little more suitable for yes/no results,” says Van der Veer. “But that’s typically the question with infectious diseases, do you have it or not?” He explains that besides for infectious diseases, PCR is also used for other kinds of testing. For example, when you want to measure the concentration of certain DNA/RNA which is normal in low concentrations, but may indicate cancer when above a specific limit. “PCR can more accurately measure the concentration of RNA or DNA in a sample. So, if that’s your question, PCR is better.” But, he adds, “that usually doesn’t concern infectious diseases.”
Vision for the future
“We’re also developing this test for malaria,” says Van der Veer. “Malaria is particularly prevalent in rural areas of African countries, where you don’t have the medical infrastructure with laboratories that we have here.” In addition, he adds, it is very important that medication is only taken when someone actually has malaria. This is because unnecessary use of medication creates resistance to it among the population. “That means you need quick and inexpensive diagnostics for someone suspected of having malaria so that you immediately know if it’s really malaria,” he continues. “And this method is ideally suited for that. Because it’s inexpensive, quick, and easy.”
In addition to Covid-19 and malaria, Merkx and Van der Veer’s technique can in principle be used for any infectious disease. This way, with a little bit of help from the Oplophorus gracilirostris, they hope to provide you with your results as quickly as possible.



Discussion